Part:BBa_K627015
Plasmid backbone pSB1A3 carrying the lac operon induction system
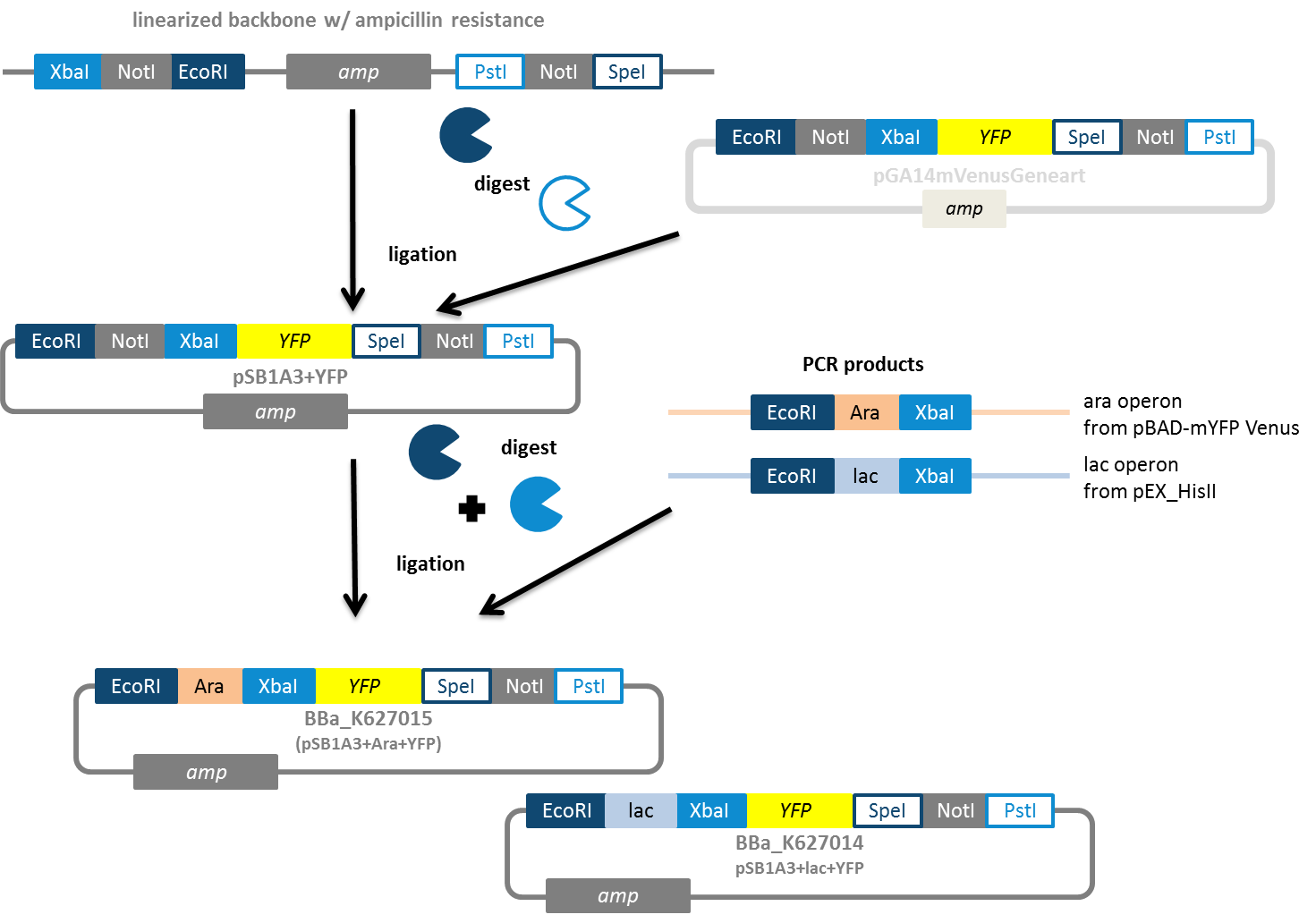
In a subtask of our work we wanted to construct auxiliary expression backbones with inducible promotors. In contrast to constitutive systems inducible systems only express the protein of interest after adding an inducer, which can be for example AHL or arabinose. We decided to construct IPTG- and arabinose-inducible systems. Therefore we amplified the promotor region and fused it to a reporter gene, which is constituted by YFP. Using this we have the option to verify the presence of the inducible promotor and also the function of the induction process by fluorescence. Our idea was to use the reporter gene as a placeholder. Via restriction enzyme digestion using the iGEM restriction enzyme site you will be able to replace the reporter gene with your gene of interest. As vector backbone we made use of the pSB1A3, which has ampicillin resistance (Fig. 8). The constructs using pSB1K3 (kanamycin resistance) and pSB1C3 (chloramphenicol resistance) are still on the anvil.
We successfully tested the IPTG- and arabinose-inducible system (Fig 9). Using fluorescence microscopy we were able to detect the YFP-expression after an induction time of approximately 1.5 hrs. To prove the hypothesis we opposed an induced sample with a non-induced control for each promotor. By use of brightfield combined with differential interference contrast microscopy we traced the E. coli cells and switched then to the YFP detecting channel to investigate the fluorescence of these cells.
We also tested the inducible systems by using fluorescence spectroscopy. For this experiment we induced both systems at a time when the cultures were located in phase of exponential growth. After inducing, the cultures were analyzed by fluorescence spectroscopy. In this analyze the cells were excitated by 500 nm. The resultant emission was measured in a spectrum between 510 and 580 nm. (Fig 10;11)
In contrast to the control of the arabinose inducible system the Lac-control also shows fluorescence. This demonstrates the leaky Lac-promotor, what is also a scientifically proven fact. A leaky promotor means that the promotor is not regulated in a leakproof manner. Even in repressed conditions the transcript can arise.
The lacking of the Lac-I gene on the vector is the reason for expression of YFP in both controls. For further research in this field it is necessary to clone the Lac-I gene inside the constructed vector.
Source
- pGA14mVenus Geneart
- Linearized BioBrick backbones
Design Notes:
We decided to construct IPTG- and arabinose-inducible systems. Therefore we amplified the promotor region and fused it to a reporter gene, which is constituted by YFP. Using this we have the option to verify the presence of the inducible promotor and also the function of the induction process by fluorescence. Our idea was to use the reporter gene as a placeholder. Via restriction enzyme digestion using the iGEM restriction enzyme site you will be able to replace the reporter gene with your gene of interest. As vector backbone we made use of the pSB1A3, which has ampicillin resistance (Fig. 8). The constructs using pSB1K3 (kanamycin resistance) and pSB1C3 (chloramphenicol resistance) are still on the anvil.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix. - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
| None |